Hi, I'm Shiwani Limbu
A
Self-motivated and highly enthusiastic Bioinformatician with experience in molecular biology and cell culture techniques. Demonstrates collaborative behavior, with a high degree of integrity, accuracy, and attention to detail.
About
I am a Bioinformatics Graduate Student at University of California, Merced. I enjoy solving computational biology research problems, programming, and effective communication. Always strive to bring 100% to the work I do. I have worked on technologies like Python, R, Bulk and Single cell RNAseq, Chipseq, and ATACseq during my Master's. I have three years of computational biology experience, which has helped me strengthen my experience in programming and data analysis. I am passionate about developing useful workflows and applications that can help bench scientist gain deeper insights into their experimental data.
- Programming Languages: Python, R, Bash
- Libraries: NumPy, Pandas, Scikit-learn
- Frameworks: TensorFlow
- Tools: Git, Docker, Slurm
- Statistical Modelling: Hypothesis testing, Linear regression, and Dimensionality reduction using Principal Component analysis.
- Bioinformatics Methods: GATK, Bowtie2, Hisat2, samtools, fastqc, picard, featureCounts, IGV, DESeq2, limma, MuTect2, SRAToolkit, Seurat, Scanpy, UCSC Genome browser, Ingenuity Pathway Analysis, CLC Genomics
- Wetlab Technologies: Flow cytometry, Stem cell differentiation, Cell culture (Stem cells, HUVECS, Embryonic stem cells, Fibroblast, Smooth muscle cell, A549 LUAD cell line, and Pericytes) maintenance and freezing, Tumor spheroid generation, Transwell migration assay, 3D cells culture, Cell staining, Western blot, PCR, Gel electrophoresis, Plasmid purification, cell culture imaging, Fiji image analysis and image processing.
Looking for an opportunity to work as a Bioinformatics Research Associate/Analyst where I can utilize my computational biology skills and wet lab experience to design computational methodologies and workflows that can enhance research outcomes.
Experience
- Applied NGS RNASeq data analysis using Bowtie2, Hisat2, samtools, and featurecounts, and MuTect2 for somatic variant call.
- Led multiomics data analysis (RNAseq, copy number variant, methylation, and Chipseq) to identify genomic changes associated with immune cell infiltration in Cancer.
- Applied Scanpy and Seurat to analyze Single-cell RNAseq data.
- Applied MACS2 and SEACR for CUT&TAG peak calling.
- Developed a python package (VariantAnnotator and TMAP).
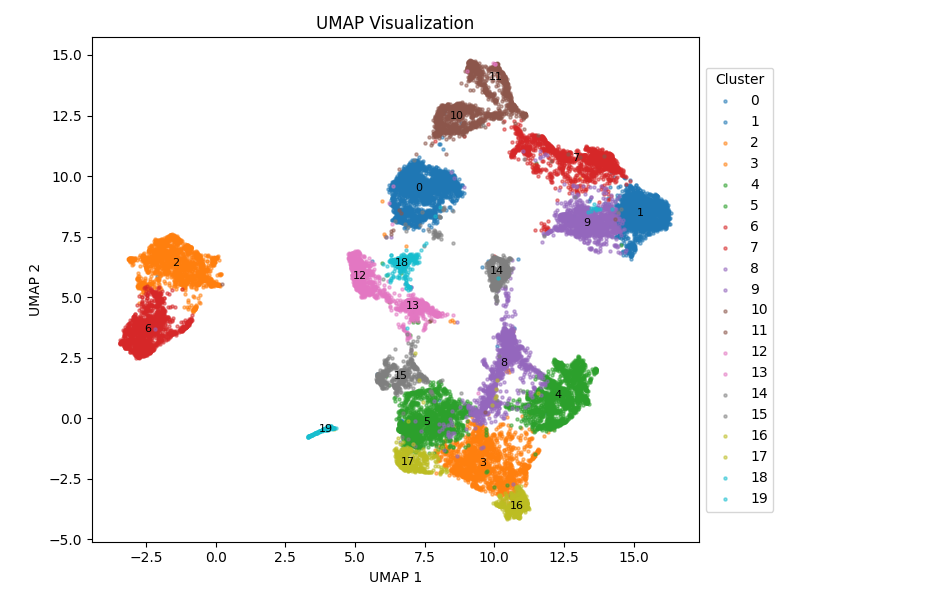
- Applied differential expression, immune cell infiltration, gene set enrichment analysis (GSEA), and survival analysis to identify Lung Adenocarcinoma biomarker .
- Developed machine learning model to differentiate between endothelial and non-endothelial cells using Transcriptomic data.
- Experienced in differentiating human embryonic stem cells (hESCs) into mature endothelial cells to advance research in vascularization within the Tumor Microenvironment.
- Generated Tumor spheroids and implemented protocols for testing cell migration.
- Used Flow cytometry for cell sorting, as well as antibody titration.
- Skilled in using Ultracentrifuge for organelle isolation and protein separation.
- Proficient in using Spectrophotometers for quantification of bacterial growth and enzyme activity.
- Proficiently utilized Thermal Cycler and Gel Electrophoresis in PCR experiments for DNA amplification and analyzing gene repeat size variations through electrophoretic DNA separation.
- Plasmid Isolation and DNA Quantification, successfully isolated pGlo plasmid from overnight cultures of transformed cells and determined plasmid DNA concentrations in E. coli samples.
- Applied Affinity Chromatography for purification of GFP from E. coli cells transformed with pGLO™.
- One year of experience working as a lab safety manager.
Bioinformatics Projects
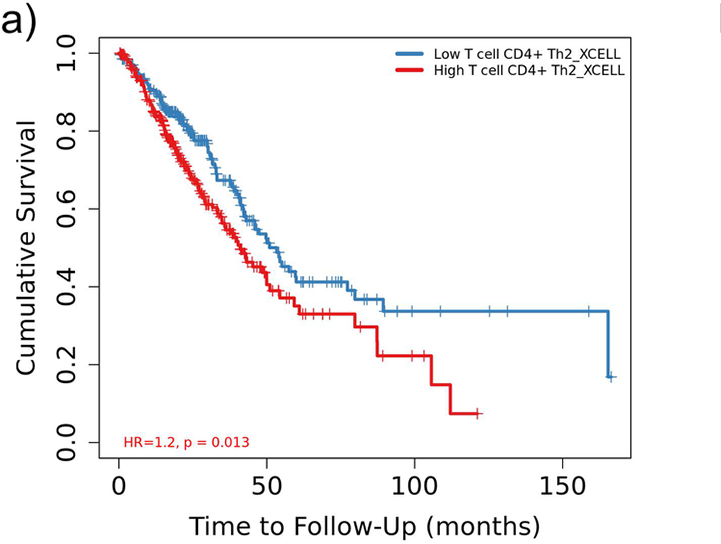
Detecting clinicopathological parameters and prognosis in lung adenocarcinoma
Skills
Languages and Databases
 Python
Python
 R
R
 Shell Scripting
Shell Scripting
Libraries
 NumPy
NumPy
 Pandas
Pandas
 scikit-learn
scikit-learn
 matplotlib
matplotlib
Technologies
 Git
Git
 Slurm
Slurm
 Docker
Docker
Education
University of California, Merced
Merced, CA, USA
Degree: Master of Science in Quantitative and Systems Biology
CGPA: 3.93/4.0
- Bioinformatics
- Epigenetics & Gene Expression
- Molecular Immunology
Relevant Courseworks:
Dehradun, India
Degree: Bachelor of Technology in Biotechnology
CGPA: 3.4/4
- Programming in C
- Advance Molecular Biology
- Bioinformatics and Biostatistics
Relevant Courseworks: